Large Language Models for Clinical Applications: Part 1
Insights and considerations about the clinical applicability of LLMs, and the importance of domain knowledge in guiding these systems
Here’s a (very complementary 😀) TL;DR of this post, generated by GPT-4!
I've been involved in clinical Natural Language Processing research for the past eight years -- first as a Computer Science student and, more recently, as a medical student. A lot of my previous research has focused on Named Entity Recognition, Information Retrieval, and automatic document generation from patient medical records. More recently, I've been exploring the applicability of Large Language Models (LLMs) like GPT-3 in the clinical context, including on the tasks I mentioned above, but also new ways of using them to interact with clinical guidelines and records. I've been building MedQA in part to explore how to harness the strengths and limit the drawbacks of LLMs in the clinical domain.
There have been many public comments and demos on the roles of LLMs in the clinical domain from a generative sense (such as writing or autocompleting medical notes). I'd like to share some insights and considerations about the clinical applicability of these models from a classification and decision-making perspective that I've discovered over the course of building MedQA, using it at the hospital, and sharing it with other fellow med students and doctors. I'd also like to motivate the importance of having and utilizing domain knowledge in guiding these systems. (There have also been several interesting technical learnings from my experiences as well that I'll focus on in a future post!).

Where LLMs can fit into clinical applications — and specifically how they can extend beyond existing Clinical AI implementations
A deeper understanding and reflection of clinical context
A common application of NLP in the clinical domain is Information Retrieval for clinical question-answering (ie. Fetch latest treatment protocols for Condition X from PubMed and answer specific questions about them; Calculate CHA₂DS₂-VASc Score for Atrial Fibrillation given an unstructured medical record). General Information Retrieval techniques — like computing embedding vector similarity, other transformer-based techniques, and even classical techniques like Okapi BM25 — have been used in clinical systems with a reasonable degree of success (more about this in my Master’s thesis, if interested). However, understanding query intent in a flexible and extensible manner and reflecting that in the results is something that is easy for trained physicians to understand and difficult for most NLP systems to do.
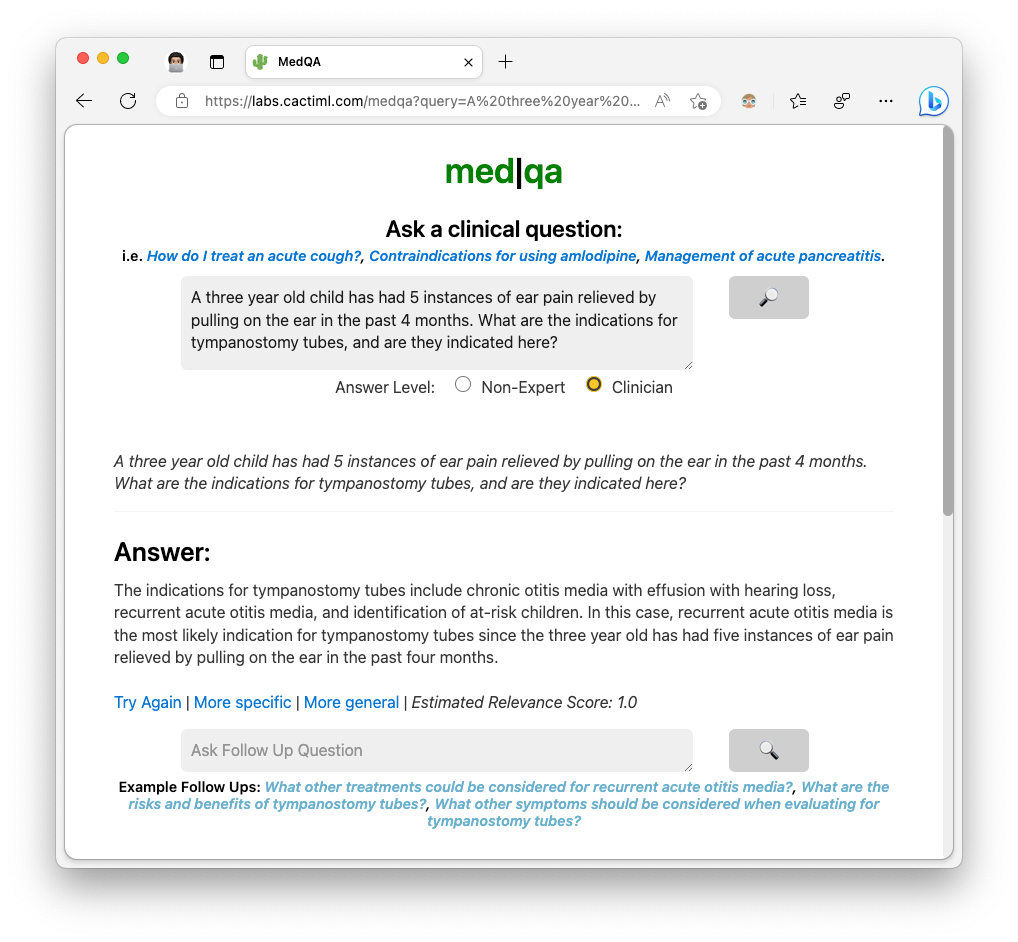
Some examples I’ve come across through observing how people use MedQA: MedQA is a reference tool that can answer clinical questions and follow-up questions in natural language, along with references to sources. With each query, there is important clinical context associated with the query representing the user intent. Some queries represent a simple lookup of facts. However, there are more complex use-cases, such as:
requesting information about differential diagnoses — therefore, the system should do clinical reasoning in a Bayesian-like manner given background evidence
wanting to compare and contrast various conditions or approaches — the system needs to identify what the metrics to make these comparisons/contrasts are, which also may be influenced by background evidence
varying layers of complexity of a response, such as a concise step-by-step list of a surgical procedure vs. detailed epidemiology of a condition — the system needs to be able to reflect these desires
While previous deep learning systems could handle one or more of these use-cases, the ability for LLMs like GPT to interpret and act on clinical context either, a) out-of-the-box; or b) through explanation in a prompt, offers a degree of generalizability and flexibility that is necessary for broad integration into clinical use-cases.
To illustrate, through these methods, MedQA can:
Detect if a query lends itself well for a table-based response, and respond accordingly
Identify whether the query represents a desire for a succinct, step-by-step description or a more elaborate description
Elaborate or be more concise as needed
Enabling “Fuzzy” Clinical Decision-Making Algorithms
Many clinical algorithms (whether implemented as a machine learning model or as part of hospital policy) rely on categorizing and compartmentalizing information. Things like how a patient is presenting, their past medical history, and how a patient is feeling are often reduced to checkboxes that are then used for further decision-making. This is also present on a less-explicit level: I have previously worked on several projects applying clinical NLP techniques to hospital workflows involving patient notes, and a big part of the technical stack involved the use of neural networks to perform entity extraction from the unstructured text. These extracted entities would then be used for downstream tasks.

However, particularly in medicine, factors like patient history can contain significant nuance and complexity that is important for decision-making but not able to be captured using reductive techniques. In undergrad, I was on a research team participating in the National NLP Clinical Challenges (n2c2) for cohort identification, where the challenge was classifying whether or not particular patients’ medical records reflected eligibility for each of 13 different clinical trials. We ran into this same issue back then — there are so many nuanced ways a criteria like “patient presents under the influence of a substance” can present in a medical note that training a single, specialized binary classifier for each of the 13 subtasks was not time-efficient (and, moving beyond a research task, scalable). We ultimately ended up performing very well with a amalgamation of rule- and ML-based subsystems, but it is obvious how such a time-intensive and narrow approach is not suitable for widespread clinical use-cases.
One ability of LLMs that I've seen reflected when using MedQA or ChatGPT which I'm excited to further explore is the ability to capture and act on data that is nuanced and difficult to formally reduce. Being able to encode more "fuzziness" into medical systems and moving away from rigid, inflexible inputs to algorithms, can bring tools that can augment clinical decision-making one step closer to performing in a more natural, comprehensive manner necessary for optimal health outcomes.
A case for clinical domain knowledge in building with LLMs
While ChatGPT has been very helpful for speeding up my personal development in technical domains I am not very familiar with (such as frontend development, and building simple UIs for several small side projects), I have used a surprising degree of experience and domain knowledge that I’ve picked up through medical school when working to align GPT with specific clinical goals.
One such area was in prompt construction — I have gradually built up a relatively lengthy prompt of 300+ words that encompasses a significant portion of MedQA functionality. Some interesting areas where I used experiences from medical school:
(side note: if interested, Google’s MedPaLM paper also touches on the use of prompt engineering in the clinical domain)
Identifying user intent and responding accordingly:
As described above, different types of clinical queries call for different types of answers. A lot of the specifics come from drawing from my own experiences during clerkships in medical school, as well as feedback from doctors I shared MedQA with. For instance, during my surgery clerkship, I would quickly read a refresher of the steps of a particular surgical case. Thus, when I asked for a quick overview of a procedure, I was looking for a high-level description of the surgery, along with information like indications and complications. On the other hand, if I was inquiring about a potential diagnosis, an accompanying reasoning for the diagnosis and other differential diagnoses would be helpful.
Most of the degree of differentiation in output based off these criterion is currently done through detailed prompt crafting. Interestingly, I thought back to the early days of medical school, when we were being taught how to give effective clinical presentations and the sort of information to include, to inform my prompt 😊 . There is also a large role for integrating domain knowledge into the information retrieval phase of LLM pipelines as well.
Identifying areas when to use (and when not to use) LLMs
We went through many areas where LLMs can shine in the clinical context — doing some level of reasoning (at least, something that looks like reasoning and is close enough for many contexts); being able to synthesize topics and manipulate them to compare/contrast, simplify, highlight components from it; capturing a great degree of nuance from questions or contexts. However, for some clinical queries or sub-questions, LLMs are not the right answer. Examples include drug dosage lookup, trying to identify specific numerical criterion specified by guidelines, or any other sort of highly objective and structured piece of information. For these use-cases, utilizing a hybrid approach of fetching that data independently and ingesting it into the pipeline, is the correct solution.
We can see this playing out with the launch of ChatGPT Plugins (although I am still on the waitlist so don’t have any firsthand experience), but currently, domain knowledge offers necessary insight into how to structure applications to properly leverage LLMs. Several features of MedQA — including if there are medication or clinical algorithm-based information that needs to be pulled from elsewhere, detecting whether a user is asking a clinical question vs. trying to answer a multiple choice question (not the focus of the app!), or offering to return the information as a table — come from understanding the strengths and limitations of LLMs in this context.
I originally built MedQA as a vehicle for exploring how to use LLMs in the clinical context, from both a domain-specific as well as technical perspective, to help guide my future projects. In addition to clinical insights (more of which I hope to share in a “Part 2” of this post), I’ve also gained some valuable experience on the technical side of things that have helped me tremendously in building around LLMs.
If you’re interested in any of my other (clinical- or non-clinical) work and projects, please check out my website (https://samrawal.com/) or follow me on Twitter (@samarthrawal). Thanks for reading!